For details of the systems used, see the corresponding pages for NAMD v.2.6, or view the scripts included at the end of this page. The ApoA1 benchmark is as distributed by the NAMD developers. If you'd rather prefer cutting a long story short, see the timings (in ns/day) shown on the very last table of this page.
The following executables have been tested
Prebuilt NAMD_2.7b1_Linux-x86_64
Prebuilt NAMD_2.7b1_Linux-x86_64-TCP
-
-
For comparison, results from NAMD2.6 have also been included
For all executables (except MPI), a slurm script in the spirit of the following was used:
#!/bin/tcsh -f
/usr/local/namdtest/charmrun /usr/local/namdtest/namd2 +p16 +giga equi.namd >& LOG
exit
There is one exception though: for jobs to be run on 4 (or less) cores full advantage of the SMP capabilities was achieved by submitting without charmrun:
#!/bin/tcsh -f
/usr/local/namdtest/namd2 +p4 equi.namd >& LOG
exit
The MPI executable was the worst of all. It was so bad, that it will not be examined any further. The results from the TCP version were also systematically worse, so they will also be excluded. The results for the three remaining executables and the various systems are (in nanoseconds per day):
| | NAMD v.2.6 | Prebuilt UDP | Source UDP-SMP |
| | | | |
| 100K atoms, _8 cores | 0.78 | 0.87 | 1.00 |
| 100K atoms, 16 cores | 1.23 | 1.35 | 1.64 |
| 100K atoms, 32 cores | 1.69 | 1.72 | 2.00 |
| | | | |
| 60K atoms, _8 cores | 1.24 | 1.42 | 1.68 |
| 60K atoms, 16 cores | 1.91 | 2.00 | 2.60 |
| 60K atoms, 32 cores | | | 2.59 |
| | | | |
| 25K atoms, _4 cores | 1.70 | 1.94 | 2.53 |
| 25K atoms, _8 cores | 2.38 | 2.85 | 4.10 |
| 25K atoms, 16 cores | | | 4.04 |
| | | | |
| 1.6K atoms, _1 cores | 11.50 | 14.10 | 13.50 |
| 1.6K atoms, _2 cores | 11.80 | 13.80 | 20.60 |
| 1.6K atoms, _4 cores | | | 47.60 |
| | | | |
| 0.9K atoms, _1 cores | 22.20 | 26.70 | 25.90 |
| 0.9K atoms, _2 cores | 21.20 | 25.60 | 40.00 |
| 0.9K atoms, _4 cores | | | 79.00 |
For the udp-smp-icc executable, the results from the standard ApoA1 NAMD benchmark are:
| Number of cores | 1 | 2 | 4 | 8 | 16 | 32 |
| | | | | | | |
| Nanosecond per day | 0.097 | 0.19 | 0.37 | 0.72 | 1.38 | 2.31 |
| | | | | | | |
| Days per nanosecond | 10.325 | 5.257 | 2.730 | 1.380 | 0.723 | 0.433 |
| Parallel Efficiency (%) | - | 98.2 | 94.5 | 93.5 | 89.2 | 74.5 |
| Days/nsec for 100% efficiency | 10.325 | 5.163 | 2.581 | 1.290 | 0.645 | 0.327 |
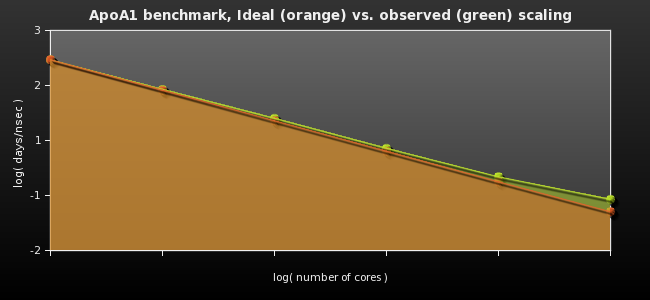
Finally, running the udp-smp-icc NAMD executable with +max_dgram_size 5800 +os_buffer_size 400000 +LBPredictor +LBLoop +noAnytimeMigration +asynciooff +setcpuaffinity +LBSameCpus +giga gave the following measurements in nanoseconds per day (compare with tables shown above):
| Number of nodes (cores) | 1 (4) | 2 (8) | 4 (16) | 8 (32) |
| ApoA1 benchmark | | | | 2.42 |
| 100K atoms | | | 1.69 | 2.40 |
| 60K atoms | | 1.67 | 2.75 | 3.22 |
| 25K atoms | 2.53 | 4.13 | 5.35 | x |
| 13.3K atoms | 7.27 | 8.28 | x | x |
| 1.6K atoms | 47.6 | x | x | x |
| 0.9K atoms | 79.0 | x | x | x |
In this table, vacant entries mean 'no measurement taken', crosses mean 'measurement taken and it was worse than that obtained with half the number of cores'.
Now the crucial question: should we be happy with these numbers ? The answer is, of course, “possibly” : 2.42 nsec/day for ApoA1 translates to approximately 0.071 seconds per step, which according to this page is
15% faster than what you would get from 128 cores on the SDSC IBM BlueGene/L. To make the comparison more meaningful (and up-to-date), we also compare with this HPC advisory dated January 2009. According to the data shown on page 9 of this PDF file, 2.42 nsec/day for the ApoA1 benchmark and 32 cores is slightly better than what you would have get from that cluster if you have 10 Gbit ethernet (and not much slower if you had infiband, running at ~2.8 nsec/day). Referring to the same slide, to get the same performance as norma does using simple gigabit, you would have to use (assuming linear scaling) somewhere between 12 and 16 nodes (corresponding to somewhere between 48 and 64 cores). In summary, and if we were hard pressed to reach a conclusion, we would have said that norma's performance appears to be satisfactory.
Further evidence for the satisfactory performance of the cluster comes with a direct comparison with the results shown in the NAMD performance page as follows: We have (unashamedly) copied the graph produced by the NAMD developers, and placed on it a white asterisk corresponding to norma's performance. The thus modified graph is the following:
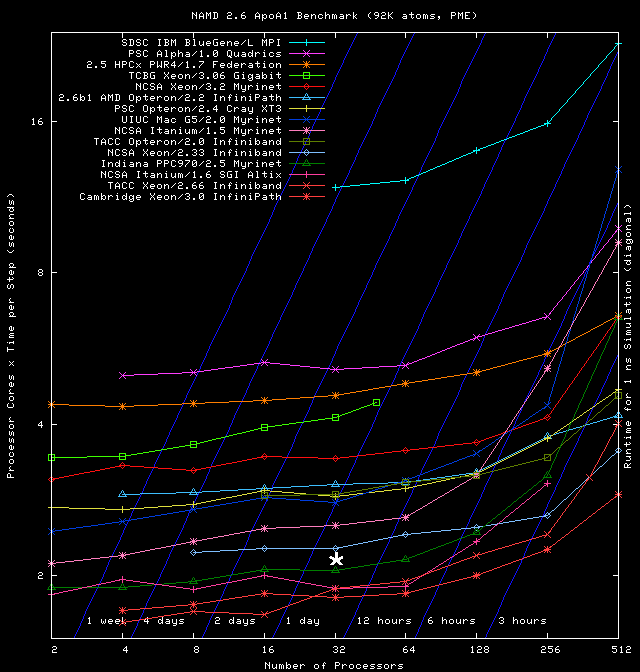
which places norma half-way between the “NCSA Xeon/2.33 Infiband” and the “Indiana PPC970/2.5 Myrinet” clusters [but do keep in mind the important difference in NAMD version (2.6 vs. 2.7b1)].
For some further tests with this version of NAMD, see here.
NAMD script for 0.9K-atoms tests
#
# Input files
#
structure ionized.psf
coordinates heat_out.coor
velocities heat_out.vel
extendedSystem heat_out.xsc
parameters par_all27_prot_na.inp
paraTypeCharmm on
#
# Output files & writing frequency for DCD
# and restart files
#
outputname output/equi_out
binaryoutput off
restartname output/restart
restartfreq 1000
binaryrestart yes
dcdFile output/equi_out.dcd
dcdFreq 200
DCDunitcell on
#
# Frequencies for logs and the xst file
#
outputEnergies 40
outputTiming 400
xstFreq 400
#
# Timestep & friends
#
timestep 2.0
stepsPerCycle 20
nonBondedFreq 2
fullElectFrequency 4
#
# Simulation space partitioning
#
switching on
switchDist 8
cutoff 9
pairlistdist 10
#
# Basic dynamics
#
COMmotion no
dielectric 1.0
exclude scaled1-4
1-4scaling 1.0
rigidbonds all
#
# Particle Mesh Ewald parameters.
#
Pme on
PmeGridsizeX 27
PmeGridsizeY 27
PmeGridsizeZ 25
#
# Periodic boundary things
#
wrapWater on
wrapNearest on
wrapAll on
#
# Langevin dynamics parameters
#
langevin on
langevinDamping 1
langevinTemp 320 # <===== Check me
langevinHydrogen on
langevinPiston on
langevinPistonTarget 1.01325
langevinPistonPeriod 200
langevinPistonDecay 100
langevinPistonTemp 320 # <===== Check me
useGroupPressure yes
firsttimestep 9600 # <===== CHANGE ME
run 20000 ;# <===== CHANGE ME
NAMD script for 1.6K-atoms tests
#
# Input files
#
structure ionized.psf
coordinates heat_out.coor
velocities heat_out.vel
extendedSystem heat_out.xsc
parameters par_all27_prot_na.inp
paraTypeCharmm on
#
# Output files & writing frequency for DCD
# and restart files
#
outputname output/equi_out
binaryoutput off
restartname output/restart
restartfreq 1000
binaryrestart yes
dcdFile output/equi_out.dcd
dcdFreq 200
DCDunitcell on
#
# Frequencies for logs and the xst file
#
outputEnergies 20
outputTiming 200
xstFreq 200
#
# Timestep & friends
#
timestep 2.0
stepsPerCycle 20
nonBondedFreq 2
fullElectFrequency 4
#
# Simulation space partitioning
#
switching on
switchDist 8
cutoff 9
pairlistdist 11
#
# Basic dynamics
#
COMmotion no
dielectric 1.0
exclude scaled1-4
1-4scaling 1.0
rigidbonds all
#
# Particle Mesh Ewald parameters.
#
Pme on
PmeGridsizeX 32 # <===== CHANGE ME
PmeGridsizeY 32 # <===== CHANGE ME
PmeGridsizeZ 32 # <===== CHANGE ME
# Pmeprocessors 4
#
# Periodic boundary things
#
wrapWater on
wrapNearest on
wrapAll on
#
# Langevin dynamics parameters
#
langevin on
langevinDamping 1
langevinTemp 320 # <===== Check me
langevinHydrogen on
langevinPiston on
langevinPistonTarget 1.01325
langevinPistonPeriod 200
langevinPistonDecay 100
langevinPistonTemp 320 # <===== Check me
useGroupPressure yes
firsttimestep 9600 # <===== CHANGE ME
run 10000 ;# <===== CHANGE ME
NAMD script for 13.3K-atoms tests
#
# Input files
#
structure ionized.psf
coordinates heat_out.coor
velocities heat_out.vel
extendedSystem heat_out.xsc
parameters par_all22_prot.inp
paraTypeCharmm on
#
# Output files & writing frequency for DCD
# and restart files
#
outputname output/equi_out
binaryoutput off
restartname output/restart
restartfreq 10000
binaryrestart yes
dcdFile output/equi_out.dcd
dcdFreq 400
DCDunitcell yes
#
# Frequencies for logs and the xst file
#
outputEnergies 80
outputTiming 400
xstFreq 400
#
# Timestep & friends
#
timestep 2.0
stepsPerCycle 20
nonBondedFreq 2
fullElectFrequency 4
#
# Simulation space partitioning
#
switching on
switchDist 8
cutoff 10
pairlistdist 12
#
# Basic dynamics
#
COMmotion no
dielectric 1.0
exclude scaled1-4
1-4scaling 1.0
rigidbonds all
#
# Particle Mesh Ewald parameters.
#
PmeGridsizeX 64 # <===== CHANGE ME
PmeGridsizeY 64 # <===== CHANGE ME
PmeGridsizeZ 54 # <===== CHANGE ME
#
# Periodic boundary things
#
wrapWater on
wrapNearest on
wrapAll on
#
# Langevin dynamics parameters
#
langevin on
langevinDamping 1
langevinTemp 340 # <===== Check me
langevinHydrogen on
langevinPiston on
langevinPistonTarget 1.01325
langevinPistonPeriod 200
langevinPistonDecay 100
langevinPistonTemp 340 # <===== Check me
useGroupPressure yes
firsttimestep 26000 # <===== CHANGE ME
run 500000000 ;# <===== CHANGE ME
NAMD script for 25K-atoms tests
#
# Input files
#
structure ionized.psf
coordinates heat_out.coor
velocities heat_out.vel
extendedSystem heat_out.xsc
parameters par_all27_prot_na.inp
paraTypeCharmm on
#
# Output files & writing frequency for DCD
# and restart files
#
outputname output/equi_out
binaryoutput off
restartname output/restart
restartfreq 10000
binaryrestart yes
dcdFile output/equi_out.dcd
dcdFreq 200
DCDunitcell yes
#
# Frequencies for logs and the xst file
#
outputEnergies 40
outputTiming 200
xstFreq 200
#
# Timestep & friends
#
timestep 2.0
stepsPerCycle 20
nonBondedFreq 2
fullElectFrequency 4
#
# Simulation space partitioning
#
switching on
switchDist 9
cutoff 11
pairlistdist 13
#
# Basic dynamics
#
COMmotion no
dielectric 1.0
exclude scaled1-4
1-4scaling 1.0
rigidbonds all
#
# Particle Mesh Ewald parameters.
#
Pme on
PmeGridsizeX 128 # <===== CHANGE ME
PmeGridsizeY 54 # <===== CHANGE ME
PmeGridsizeZ 54 # <===== CHANGE ME
#
# Periodic boundary things
#
wrapWater on
wrapNearest on
wrapAll off
#
# Langevin dynamics parameters
#
langevin on
langevinDamping 1
langevinTemp 320 # <===== Check me
langevinHydrogen on
langevinPiston on
langevinPistonTarget 1.01325
langevinPistonPeriod 200
langevinPistonDecay 100
langevinPistonTemp 320 # <===== Check me
useGroupPressure yes
firsttimestep 88000 # <===== CHANGE ME
run 50000000 ;# <===== CHANGE ME
NAMD script for 60K-atoms tests
# Input files
#
structure ionized.psf
coordinates heat_out.coor
velocities heat_out.vel
extendedSystem heat_out.xsc
parameters par_all27_prot_na.inp
paraTypeCharmm on
#
# Output files & writing frequency for DCD
# and restart files
#
outputname output/equi_out
binaryoutput off
restartname output/restart
restartfreq 1000
binaryrestart yes
dcdFile output/equi_out.dcd
dcdFreq 200
DCDunitcell yes
#
# Frequencies for logs and the xst file
#
outputEnergies 20
outputTiming 200
xstFreq 200
#
# Timestep & friends
#
timestep 2.0
stepsPerCycle 20
nonBondedFreq 2
fullElectFrequency 4
#
# Simulation space partitioning
#
switching on
switchDist 10
cutoff 12
pairlistdist 14.5
#
# Basic dynamics
#
COMmotion no
dielectric 1.0
exclude scaled1-4
1-4scaling 1.0
rigidbonds all
#
# Particle Mesh Ewald parameters.
#
Pme on
PmeGridsizeX 128 # <===== CHANGE ME
PmeGridsizeY 96 # <===== CHANGE ME
PmeGridsizeZ 64 # <===== CHANGE ME
# Pmeprocessors 16
#
# Periodic boundary things
#
wrapWater on
wrapNearest on
wrapAll on
#
# Langevin dynamics parameters
#
langevin on
langevinDamping 1
langevinTemp 320 # <===== Check me
langevinHydrogen on
langevinPiston on
langevinPistonTarget 1.01325
langevinPistonPeriod 200
langevinPistonDecay 100
langevinPistonTemp 320 # <===== Check me
useGroupPressure yes
firsttimestep 120000 # <===== CHANGE ME
run 10000 ;# <===== CHANGE ME
NAMD script for 100K-atoms tests
# Input files
#
structure ionized.psf
coordinates heat_out.coor
velocities heat_out.vel
extendedSystem heat_out.xsc
parameters par_all27_prot_na.inp
paraTypeCharmm on
#
# Output files & writing frequency for DCD
# and restart files
#
outputname output/equi_out
binaryoutput off
restartname output/restart
restartfreq 1000
binaryrestart yes
dcdFile output/equi_out.dcd
dcdFreq 200
DCDunitcell on
#
# Frequencies for logs and the xst file
#
outputEnergies 20
outputTiming 200
xstFreq 200
#
# Timestep & friends
#
timestep 2.0
stepsPerCycle 20
nonBondedFreq 2
fullElectFrequency 4
#
# Simulation space partitioning
#
switching on
switchDist 10
cutoff 12
pairlistdist 13.5
#
# Basic dynamics
#
COMmotion no
dielectric 1.0
exclude scaled1-4
1-4scaling 1.0
rigidbonds all
#
# Particle Mesh Ewald parameters.
#
Pme on
PmeGridsizeX 112 # <===== CHANGE ME
PmeGridsizeY 108 # <===== CHANGE ME
PmeGridsizeZ 108 # <===== CHANGE ME
# Pmeprocessors 8
#
# Periodic boundary things
#
wrapWater on
wrapNearest on
wrapAll on
#
# Langevin dynamics parameters
#
langevin on
langevinDamping 1
langevinTemp 298 # <===== Check me
langevinHydrogen on
langevinPiston on
langevinPistonTarget 1.01325
langevinPistonPeriod 200
langevinPistonDecay 100
langevinPistonTemp 298 # <===== Check me
useGroupPressure yes
firsttimestep 26000 # <===== CHANGE ME
run 10000 ;# <===== CHANGE ME

