Comparison between the carma-derived clusters and the RMSD (crossDCD) matrix
Focus. The aim is to plot on the RMSD matrix the areas that correspond to a selected cluster obtained from carma (using either Cartesian or dihedral PCA). Proceed as follows:
- Create a clean directory. I'll use
~/tmp/for this example. - Go to the directory you perfomed the carma cPCA or dPCA analysis.
- Write down the total number of frames used for the analysis (
tail carma.dPCA.fluctuations.dat). - Copy the
carma.clusters.datfile to the new directory (cp carma.clusters.dat ~/tmp/). - Go to the directory you've run
crossDCDfrom. - Write down the dimension of the crossDCD matrix (
wc crossDCD.matrix) - Copy the crossDCD matrix to the new directory (
cp crossDCD.matrix ~/tmp/). cd ~/tmp/- Let's say you want to analyse cluster number 1 (the top cluster). Type
make_cluster_maskand answer the questions.
You are ready to finish-it off:
carma -col - < crossDCD.matrix mv carma.stdin.ps crossDCD.ps carma -min 0 - < carma.cluster_diag.mask mv carma.stdin.ps mask.ps composite mask.ps crossDCD.ps mask.ps test.ps gs test.ps composite -density 300 mask.ps crossDCD.ps mask.ps final_image.png
… giving something like this for the first cluster of my example …
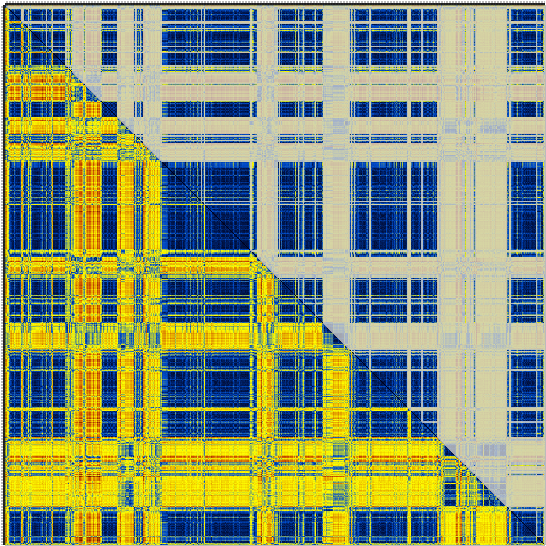
… and like this for the second …
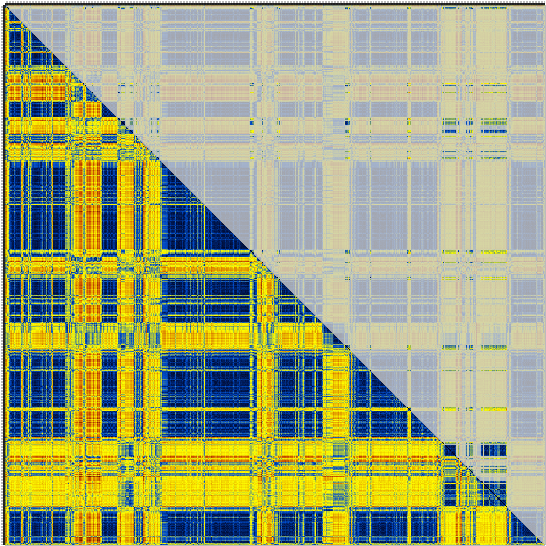
In both cases the lower half of the matrix is the original crossDCD matrix. In the upper half, everything not belonging to the cluster has been grayed-out. Enjoy.
The code for make_cluster_mask is:
#include <stdio.h> #include <math.h> #include <stdlib.h> #define MAXDIM 10000 main() { unsigned short int mat[MAXDIM][MAXDIM]; FILE *in; FILE *out; int cluster; int totframes; int i, j; char string[300]; int frame, c; int pos, dim; in = fopen( "carma.clusters.dat", "r"); if ( in == NULL ) { printf("Can't open a carma.clusters.dat file in the current directory. Abort.\n\n"); exit(1); } out = fopen( "carma.cluster.mask", "w"); if ( out == NULL ) { printf("Can't open a carma.cluster.mask file in the current directory. Abort.\n\n"); exit(1); } printf("Number of cluster to plot: "); if ( scanf("%d", &cluster) != 1 ) { printf("\nOne integer number expected. Abort.\n"); exit(1); } printf("Total number of frames of the original trajectory: "); if ( scanf("%d", &totframes) != 1 ) { printf("\nOne integer number expected. Abort.\n"); exit(1); } printf("Dimension of matrix to prepare (should be identical with CrossDCD matrix): "); if ( scanf("%d", &dim) != 1 ) { printf("\nOne integer number expected. Abort.\n"); exit(1); } if ( dim >= MAXDIM ) { printf("\nOops. Increase MAXDIM and re-compile.\n"); exit(1); } for ( i=0 ; i < dim ; i++ ) for ( j=0 ; j < dim ; j++ ) mat[i][j] = 0; while ( fgets( string, 299, in ) != NULL ) { if ( sscanf( string, "%d %d", &frame, &c ) != 2 ) { printf("Corrupted carma.clusters.dat file ? Abort.\n"); exit(1); } if ( c == cluster ) { pos = (int)((1.0 * dim * frame) / ( totframes ) + 0.50); mat[pos][pos] = 1; } } fclose(in); for ( j=0 ; j < dim ; j++ ) { if ( mat[j][j] > 0 ) { mat[j][j] = 0; pos = j; for ( i=0 ; i < dim ; i++ ) { mat[i][pos] += 1; mat[pos][i] += 1; } } } for ( i=0 ; i < dim ; i++ ) { for ( j=0 ; j < dim ; j++ ) if ( mat[i][j] > 1 ) fprintf(out, "1.0 " ); else fprintf(out, "0.5 " ); fprintf(out, "\n"); } fclose(out); out = fopen( "carma.cluster_diag.mask", "w"); if ( out == NULL ) { printf("Can't open a carma.cluster_diag.mask file in the current directory. Abort.\n\n"); exit(1); } for ( i=0 ; i < dim ; i++ ) { for ( j=0 ; j < dim ; j++ ) if ( i > j ) fprintf(out, "1.0 " ); else if ( mat[i][j] > 1 ) fprintf(out, "1.0 " ); else fprintf(out, "0.2 " ); fprintf(out, "\n"); } fclose(out); }




